
- #DECOY DATABASE FORMAT PEPTIDESHAKER SOFTWARE#
- #DECOY DATABASE FORMAT PEPTIDESHAKER CODE#
- #DECOY DATABASE FORMAT PEPTIDESHAKER DOWNLOAD#
However, in the near future we will continue to expand these libraries and frameworks to provide more powerful and robust analysis tools. Also some of them are no well-tested with extreme use cases, then they fail frequently.
#DECOY DATABASE FORMAT PEPTIDESHAKER SOFTWARE#
One of the known downsides is the lack of a thorough documentation in some cases, which may cause that the software cannot be easily reused. One of the reasons behind is that the development of open source software offers the potential for a more flexible technology and potentially, quicker innovation. In fact, there has been a big progress in the development of new libraries, allowing them to be folded into other applications and pipelines as reusable building blocks. Open-Source libraries have been fundamental in building new bioinformatics tools. FDRAnalysis is a Java library which enables the upload of peptide identification results from target/decoy searches carried out by three different search engines: Mascot, OMSSA and X!Tandem. MZmine2 is Java library mainly implemented for MS preprocessing purposes and also provides several data mining algorithms (principal component analysis, clustering and log-ratio analysis) to reduce the dimensionality of the data. Access to the available functionality is provided via high-level Python scripts. Then, Multiplierz and Pyteomics are frameworks to support proteomics data analytic tasks in this language. Python is not an extensively used programming language in computational proteomics, but in recent years is gaining popularity. Other tools, packages and open-source frameworks: InsilicoSpectro was developed in Perl and offers different set of functionalities, for instance protein digestion, sequence database file readers, property estimation (pI, retention time, mass) and MS fragmentation prediction. Questions about R/Bioconductor packages can be sought on the mailing list and general questions and suggestions about R/Bioc for proteomics data analysis can be posted on the Google group. For more details and general information about R and proteomics, have a look at the ‘R for proteomics’ introductory paper ( Pubmed, pre-print) and package. Many more packages as available and new additions are released on a regular basis. In terms of peptide identification, the rTANDEM package provides an interface to the popular X!Tandem search engine and mzID allows to parse mzIdentML files. The R environment is particularly well suited for exploratory data analysis, visualisation and statistical analysis of proteomicsdata. Other packages like MSnbase, MALDIquant or xcms (widely used in metabolomics) provide higher-level abstractions to facilitate data processing and analysis.
#DECOY DATABASE FORMAT PEPTIDESHAKER CODE#
Efficient low-level access to raw data in any of the PSI standards is possible through the mzR package (which used C and C++ code from the proteowizard project, see above). All systems are designed to be intuitively and user-friendly operable.The R language has also a set of dedicated packages for the analysis and interpretation of mass-spectrometry based proteomics data.
#DECOY DATABASE FORMAT PEPTIDESHAKER DOWNLOAD#
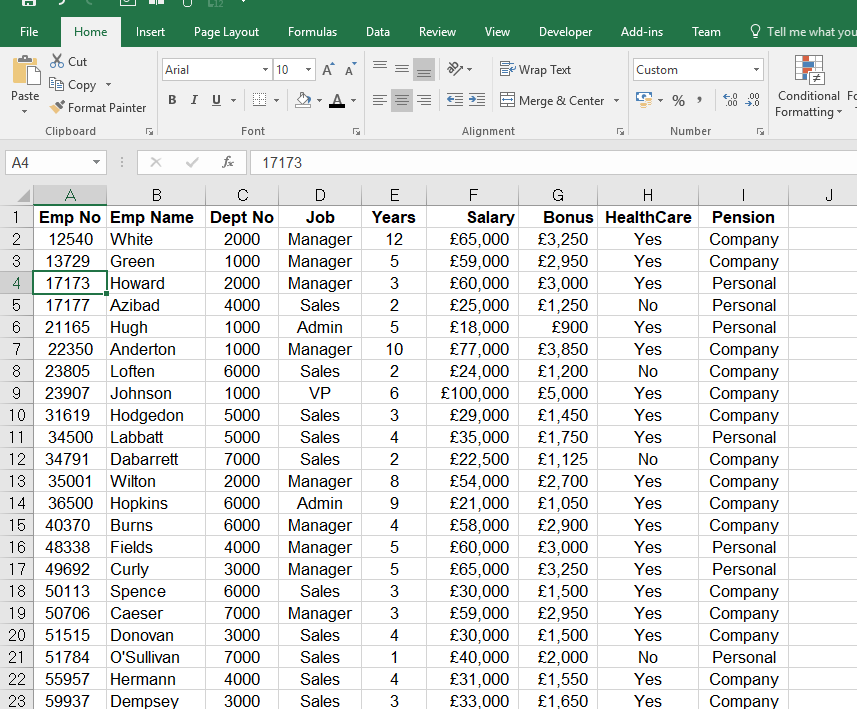
The user can easily select proteins of interest, review the according spectra and download both protein sequences and spectral library. As an additional service, we offer a web service oriented database providing all necessary high-quality and high-resolution data for starting targeted proteomics analyses. This system will be designed for high scalability and distributed computing using solutions like the Docker container system among others. In a next step, we are planning to release a web service for protein identification containing both tools. PeptideShaker combines all matches and creates a consensus list of identified proteins providing statistical confidence measures. SearchGUI is a managing tool for several search engines to find peptide spectra matches for one or more complex MS 2 measurements. In this work, we present our tools and services, which is the combination of SearchGUI and PeptideShaker. The de.NBI (German network for bioinformatics infrastructure) service center in Dortmund provides several software applications and platforms as services to meet these demands. After certain follow-up analyses, an optional targeted approach is suitable for validating the results. Typically, after capturing with a mass spectrometer, the proteins have to be identified and quantified. Computational proteomics is a constantly growing field to support end users with powerful and reliable tools for performing several computational steps within an analytics workflow for proteomics experiments.


 0 kommentar(er)
0 kommentar(er)
